Align sequences, reveal evolution
Run Clustal Omega, MAFFT, and MMseqs2 alignments instantly. Build phylogenetic trees with FastTree. No command line required.
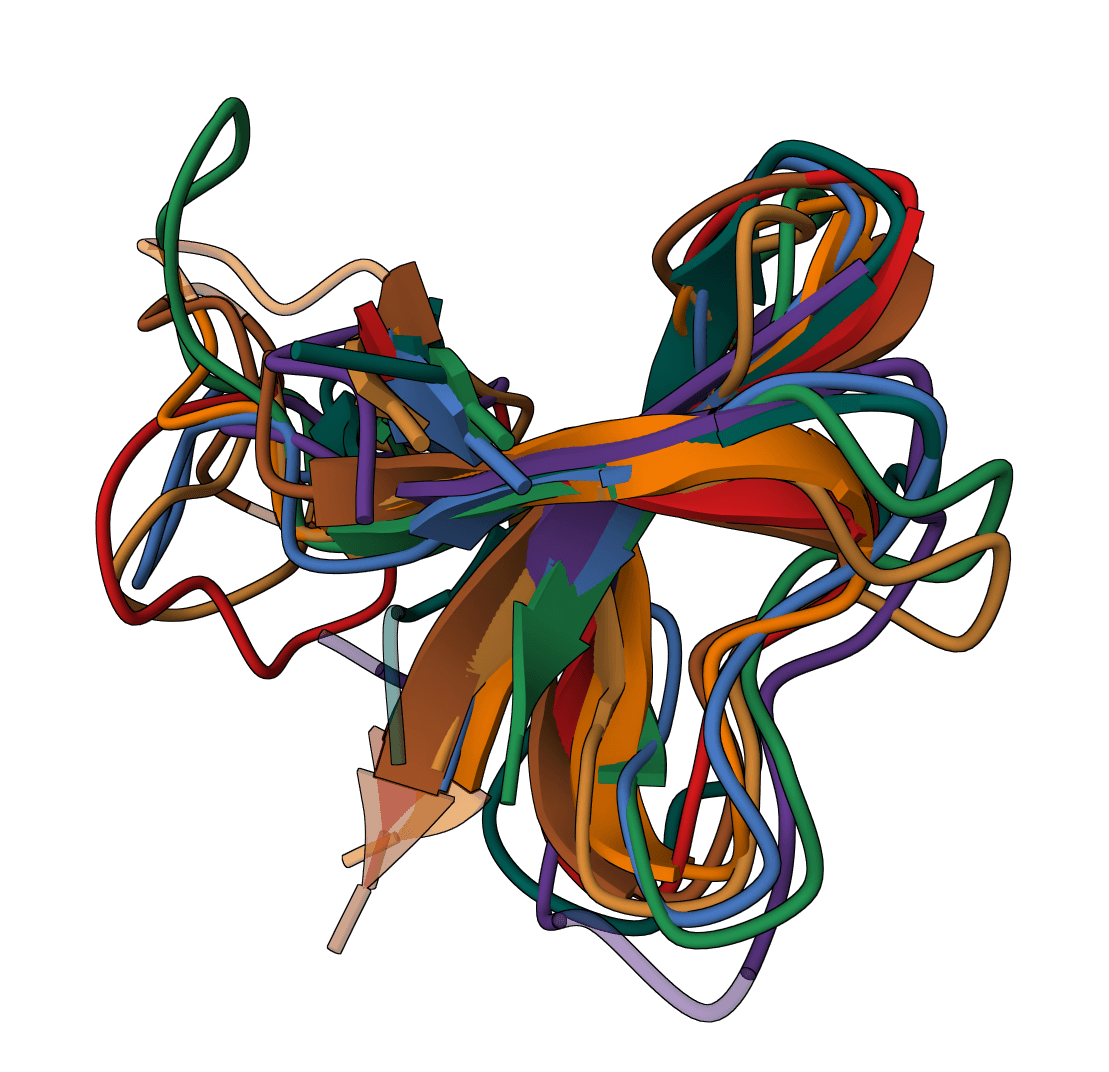
Clustal Omega
Industry-standard MSA for protein and nucleotide sequences. Handles thousands of sequences with high accuracy.
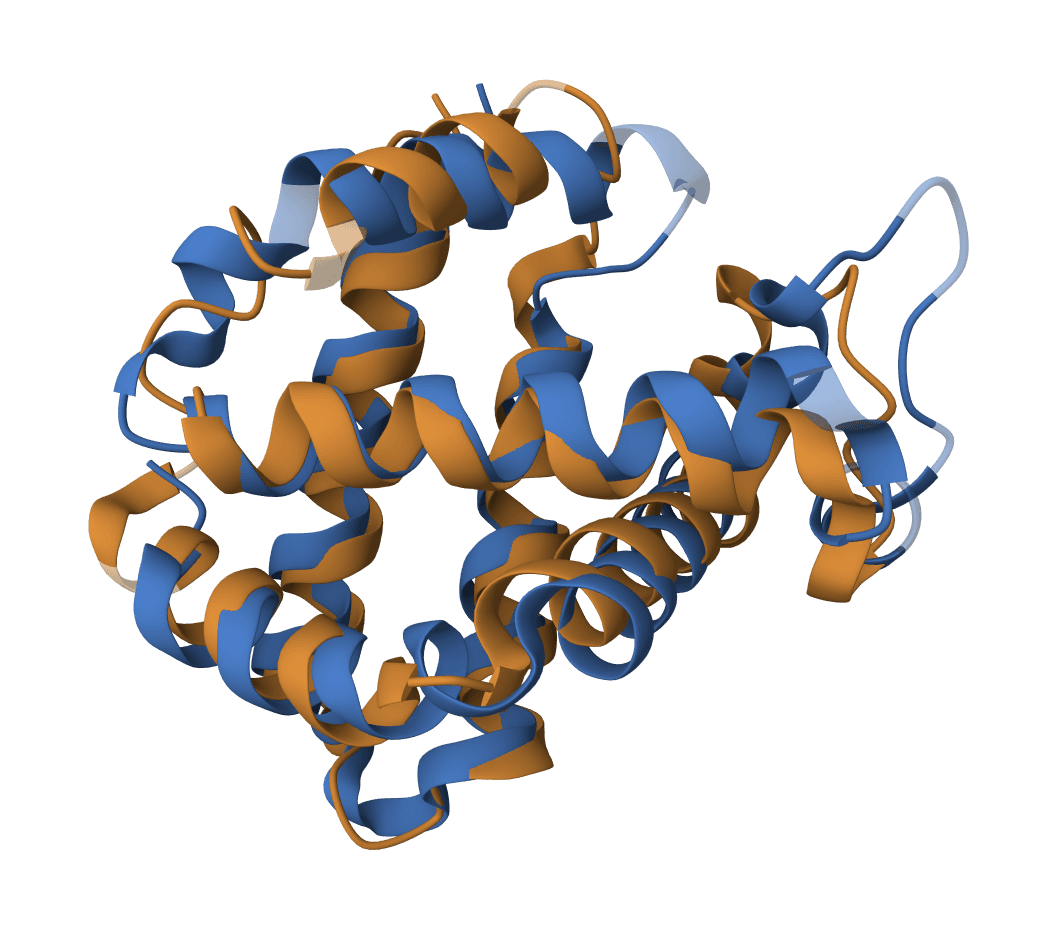
MAFFT
Fast and accurate MSA with multiple algorithms. Optimized for large-scale alignments and iterative refinement.
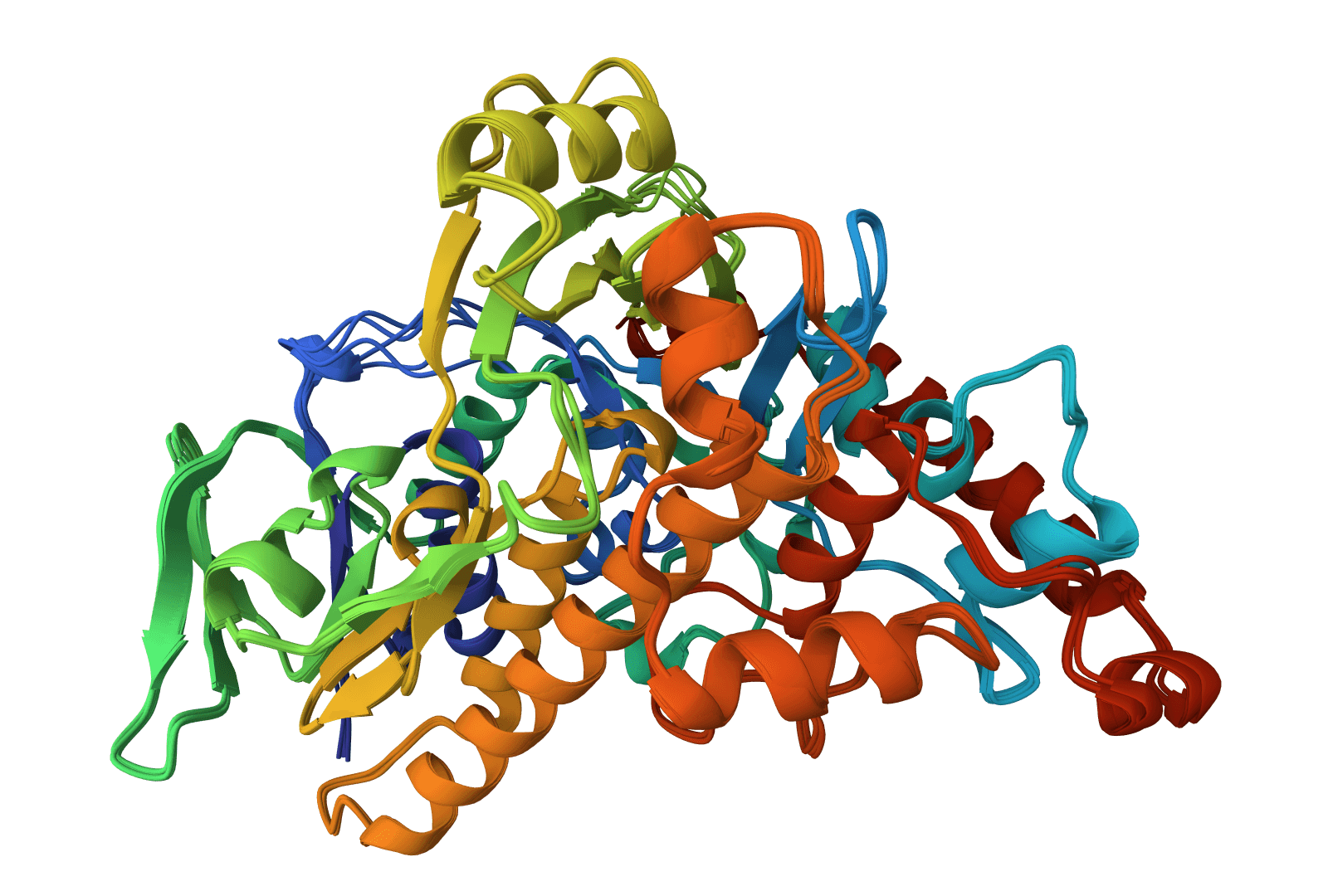
MUSCLE5
Next-generation MUSCLE with ensemble alignment. Higher accuracy than previous versions on difficult sequences.
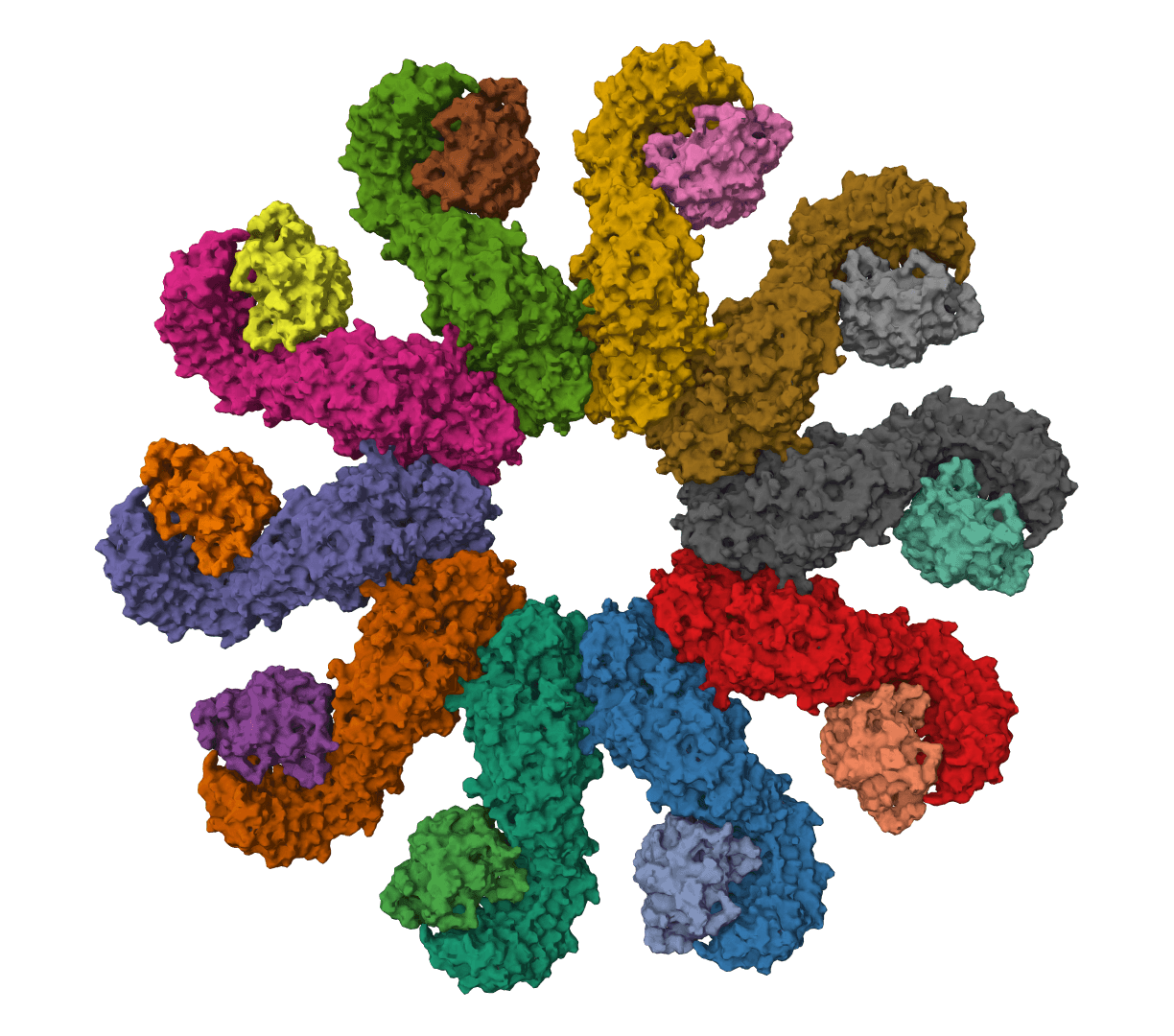
MMseqs2
Ultra-fast sequence searching and clustering. Process millions of sequences in minutes.
HMMER
Profile HMM searches for remote homology detection. Find distant evolutionary relationships.
IQ-TREE
Maximum-likelihood phylogenetics with automatic model selection. Build publication-quality trees.
Why researchers choose ProteinIQ
Run Clustal Omega and MAFFT directly in your browser—just paste sequences
Command-line alignment tools require installation and scripting expertise
Cloud compute handles large datasets with MMseqs2 clustering
Aligning thousands of sequences overwhelms local machines
Integrated workflow from alignment to tree visualization
Building phylogenetic trees requires multiple disconnected tools
Interactive alignment viewer with conservation highlighting
Interpreting raw alignment output is tedious without visualization
How it works
Professional multiple sequence alignment and phylogenetic analysis using industry-standard tools. Align protein and nucleotide sequences with Clustal Omega and MAFFT, cluster with MMseqs2, and build evolutionary trees with FastTree.
Upload sequences
Paste or upload FASTA sequences for alignment
Run alignment
Clustal Omega or MAFFT aligns your sequences
Build tree
FastTree constructs phylogenetic relationships
Visualize & export
View alignment, explore tree, download results
Related solutions
Discover complementary workflows to enhance your research.
Predict protein structures in minutes
Access AlphaFold3, Boltz, and Chai-1 models instantly—no infrastructure required, results in minutes. Design and refine structures with RFdiffusion, BoltzGen, and ProteinMPNN.
Protein design that actually works
Generate optimized protein sequences and novel binders using LigandMPNN, ProteinMPNN, SolubleMPNN, BoltzGen, and RFdiffusion—state-of-the-art deep learning and diffusion models.
Validate structures before you publish
Assess predicted and experimental protein structures using MolProbity, Ramachandran analysis, and structural comparison tools.