Screen millions, discover one
Filter compound libraries at scale with QEPPI, EvoEF2, and PocketFlow to identify promising drug candidates efficiently.
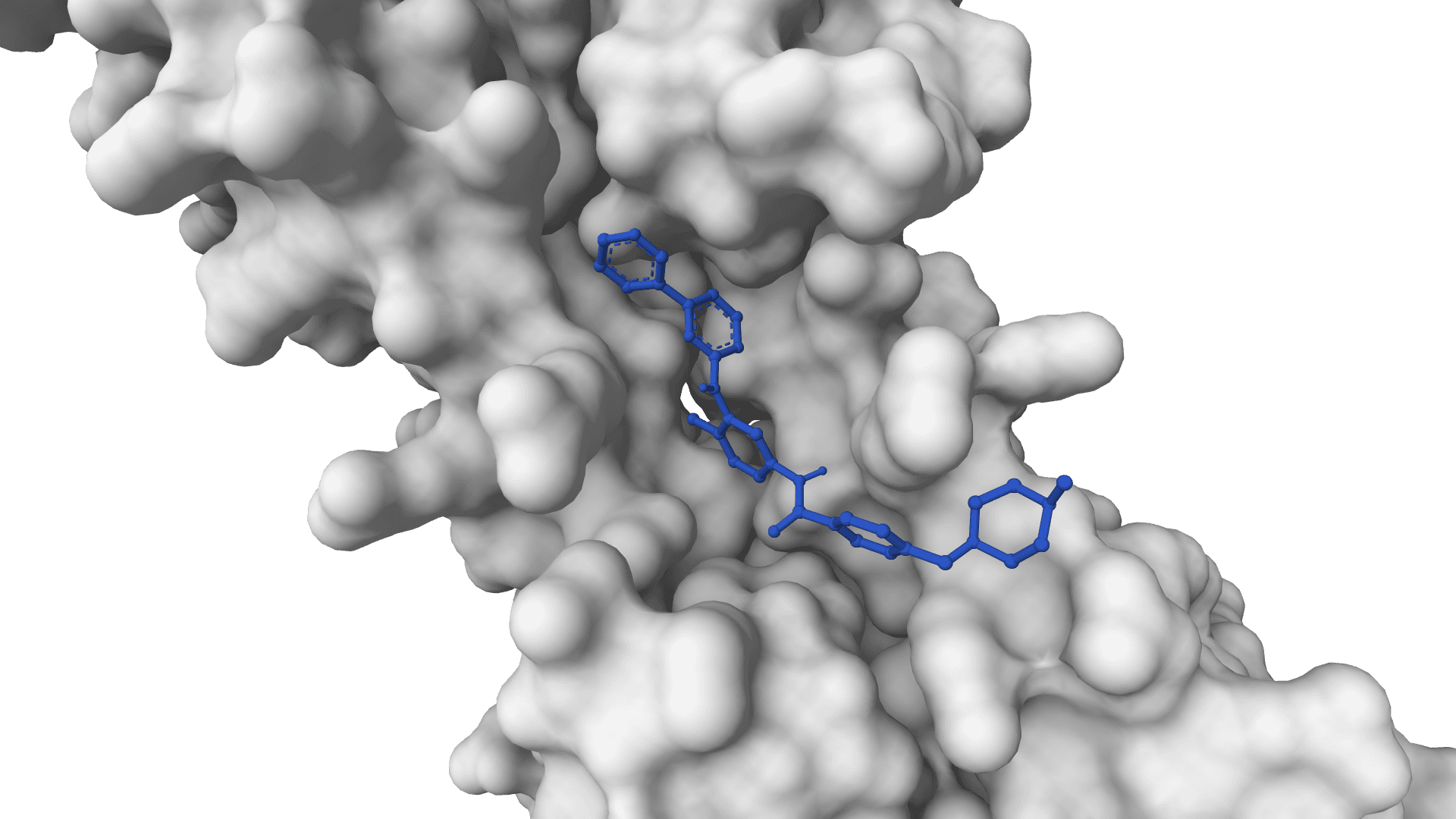
AutoDock-GPU
GPU-accelerated docking for high-throughput virtual screening. Screen thousands of compounds in minutes.
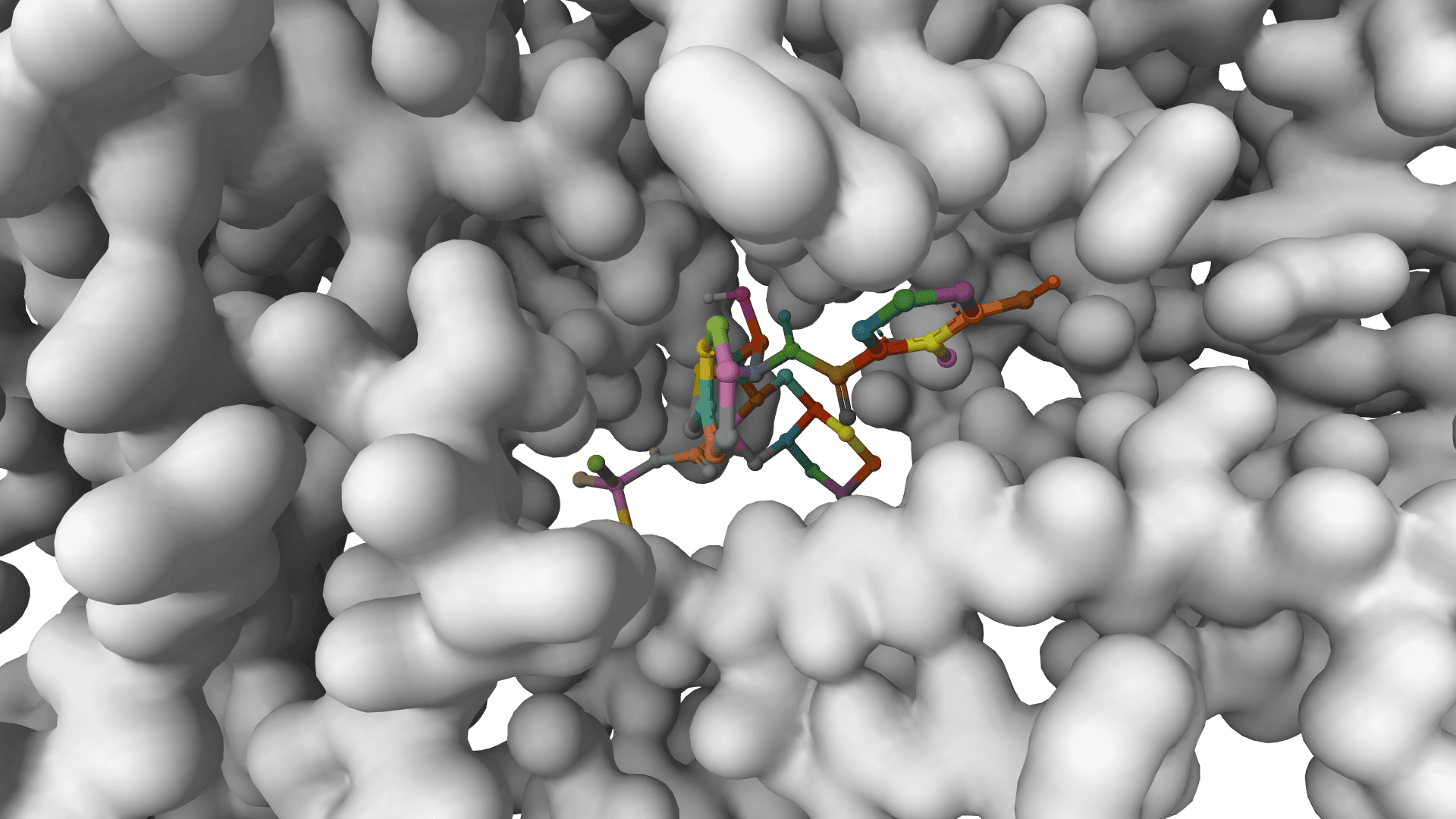
GNINA
CNN-enhanced docking with ML scoring. Higher accuracy for large-scale screening campaigns.
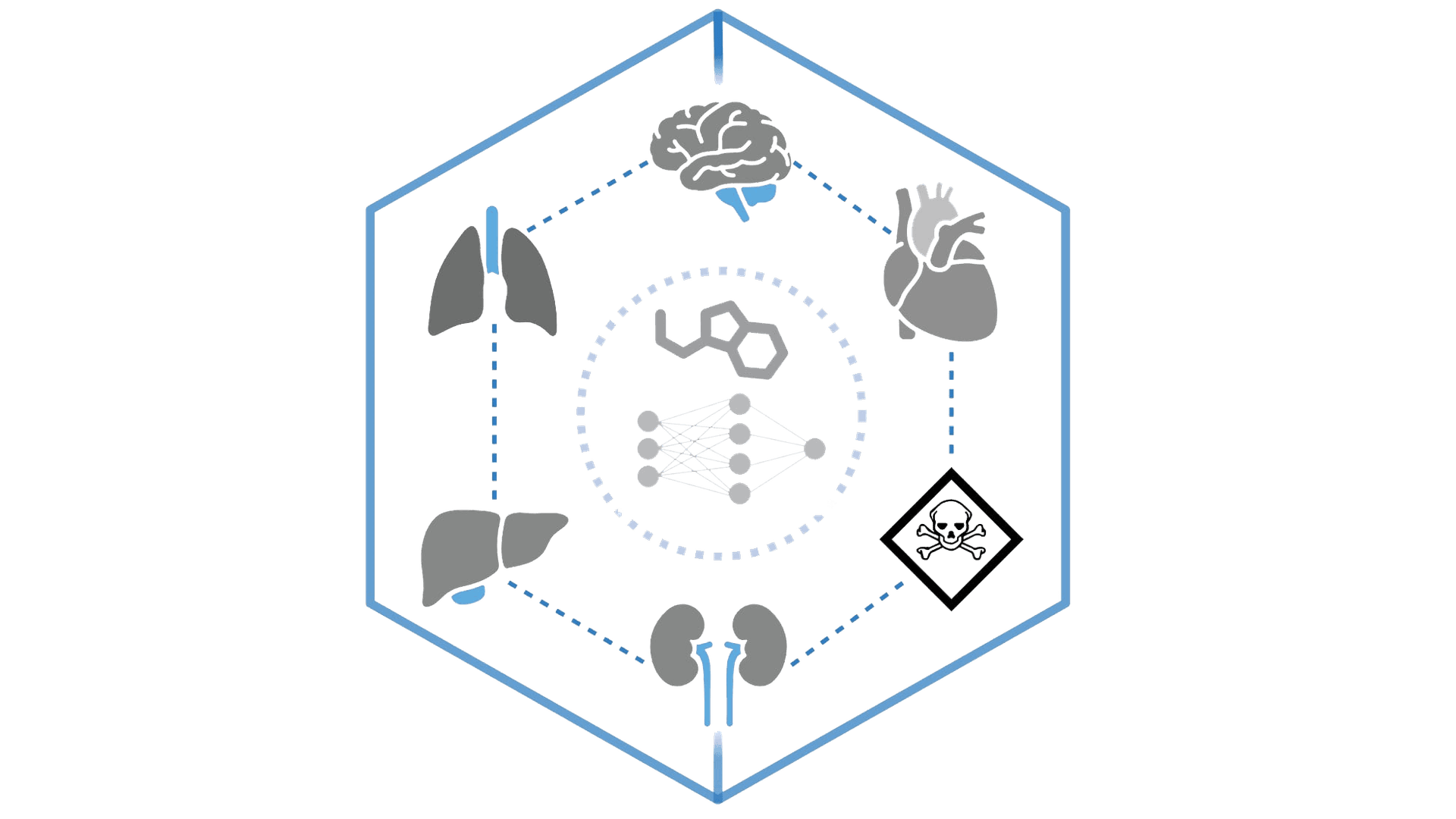
ADMET-AI
Pre-filter compounds by predicted ADMET properties. Eliminate poor candidates early in screening.

QEPPI
ML-based PPI inhibition prediction. Identify compounds that disrupt protein-protein interactions.

Lipinski's rule of 5
Filter compounds for drug-like properties. Essential pre-screening for oral bioavailability.

Molecular descriptors
Calculate 200+ molecular properties for QSAR modeling and ML-based virtual screening.
Why researchers choose ProteinIQ
Cloud-based screening scales automatically—no cluster needed
Screening millions of compounds requires dedicated HPC infrastructure
Multi-stage filtering with ML scoring improves hit quality
High false positive rates waste experimental resources
Built-in analytics and visualization for result interpretation
Managing and analyzing large result sets is overwhelming
Accessible screening tools with transparent per-job pricing
Commercial screening platforms cost tens of thousands annually
How it works
Large-scale virtual screening of compound libraries using advanced algorithms like QEPPI, EvoEF2, and PocketFlow. Identify promising drug candidates from millions of compounds.
Load library
Upload compound library or use built-in databases
Pre-filter
Apply drug-likeness and ADMET filters
Screen compounds
Run QEPPI or molecular descriptor-based screening
Rank & export
Analyze results and export top candidates
Related solutions
Discover complementary workflows to enhance your research.
Dock molecules with confidence
Screen protein-ligand and protein-protein interactions using AutoDock Vina, DiffDock, GNINA, and LightDock through a unified cloud platform.
From molecules to medicines faster
Design and validate drug candidates with integrated ADMET-AI predictions, docking simulations, and lead optimization workflows.
Predict properties before you synthesize
Calculate molecular weight, isoelectric point, extinction coefficient, stability indices, and hydrophobicity—all from sequence alone.