Protein design that actually works
Generate optimized protein sequences and novel binders using LigandMPNN, ProteinMPNN, SolubleMPNN, BoltzGen, and RFdiffusion—state-of-the-art deep learning and diffusion models.
BindCraft
End-to-end binder design pipeline combining RFdiffusion and AlphaFold2. Design high-affinity binders with experimental validation.
ProteinMPNN
State-of-the-art inverse folding with >50% sequence recovery. Design sequences for any backbone structure.
LigandMPNN
Design sequences with ligand, metal, and nucleotide context. 63.3% recovery at binding sites.
SolubleMPNN
Design soluble protein sequences optimized for expression and stability. Reduce aggregation risks.
EvoDiff
Generate diverse protein sequences using evolutionary-scale diffusion models. Unconditional and conditional generation.
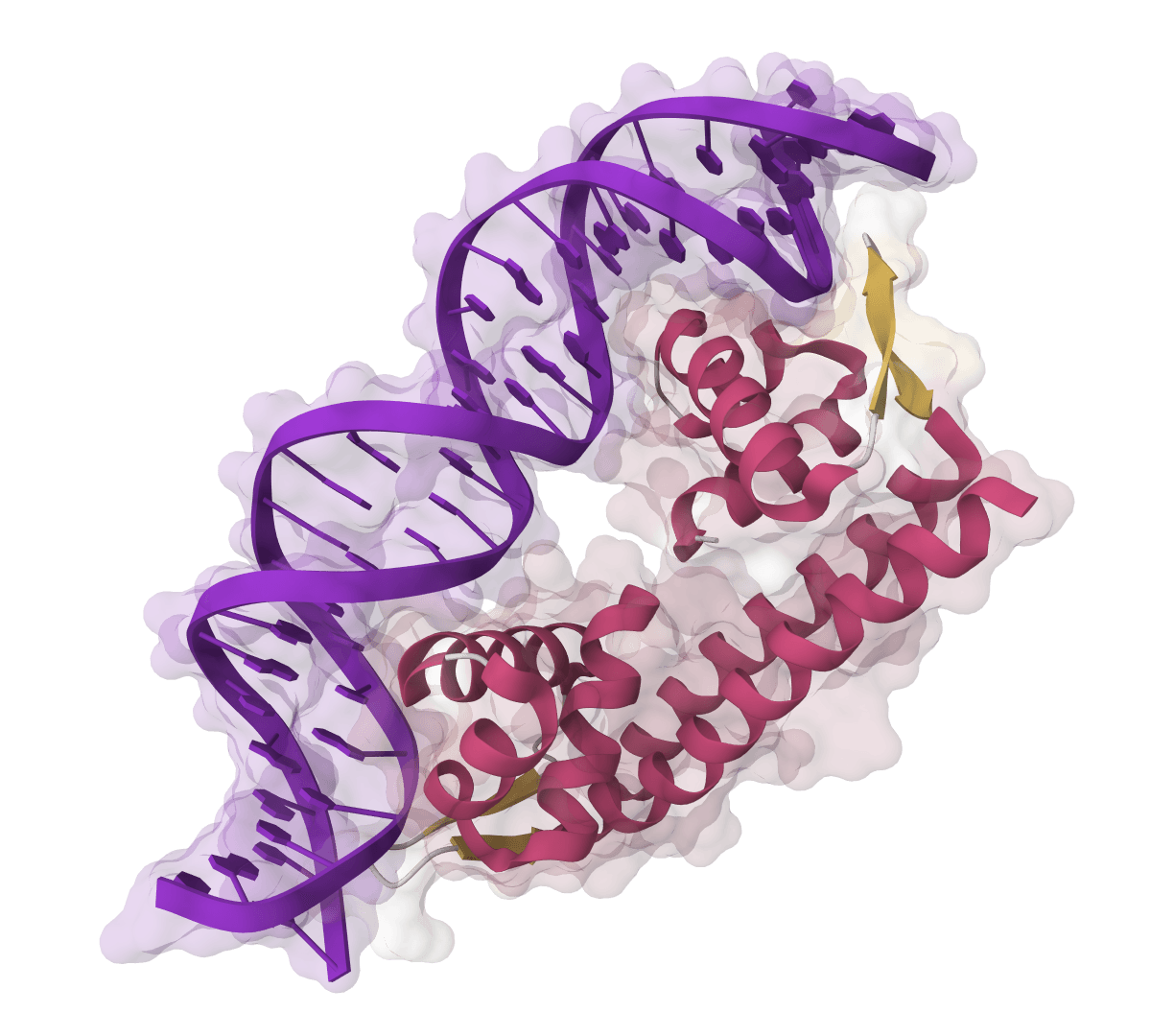
ESM-IF1
Inverse folding with ESM language models. Fast sequence design from structure with confidence scores.
Why researchers choose ProteinIQ
AI-guided design generates optimized sequences in minutes
Directed evolution requires months of experimental iteration
BoltzGen and RFdiffusion generate binders with predicted nanomolar affinity
Designing binders for novel targets is hit-or-miss
Upload structure, configure options, get sequences—no coding required
Inverse folding tools have steep learning curves
SolubleMPNN optimizes for solubility from the start
Solubility and expression issues discovered late in development
How it works
Design protein sequences and structures using cutting-edge deep learning and diffusion models. Inverse folding with LigandMPNN, ProteinMPNN, and SolubleMPNN, plus binder and structure generation with BoltzGen and RFdiffusion.
Upload structure
Provide target backbone or binding target PDB
Configure design
Set residue constraints, chain specifications
Generate sequences
ProteinMPNN or LigandMPNN designs optimized sequences
Validate & order
Check predictions and export for synthesis
Related solutions
Discover complementary workflows to enhance your research.
Predict protein structures in minutes
Access AlphaFold3, Boltz, and Chai-1 models instantly—no infrastructure required, results in minutes. Design and refine structures with RFdiffusion, BoltzGen, and ProteinMPNN.
Validate structures before you publish
Assess predicted and experimental protein structures using MolProbity, Ramachandran analysis, and structural comparison tools.
Predict properties before you synthesize
Calculate molecular weight, isoelectric point, extinction coefficient, stability indices, and hydrophobicity—all from sequence alone.