From molecules to medicines faster
Design and validate drug candidates with integrated ADMET-AI predictions, docking simulations, and lead optimization workflows.
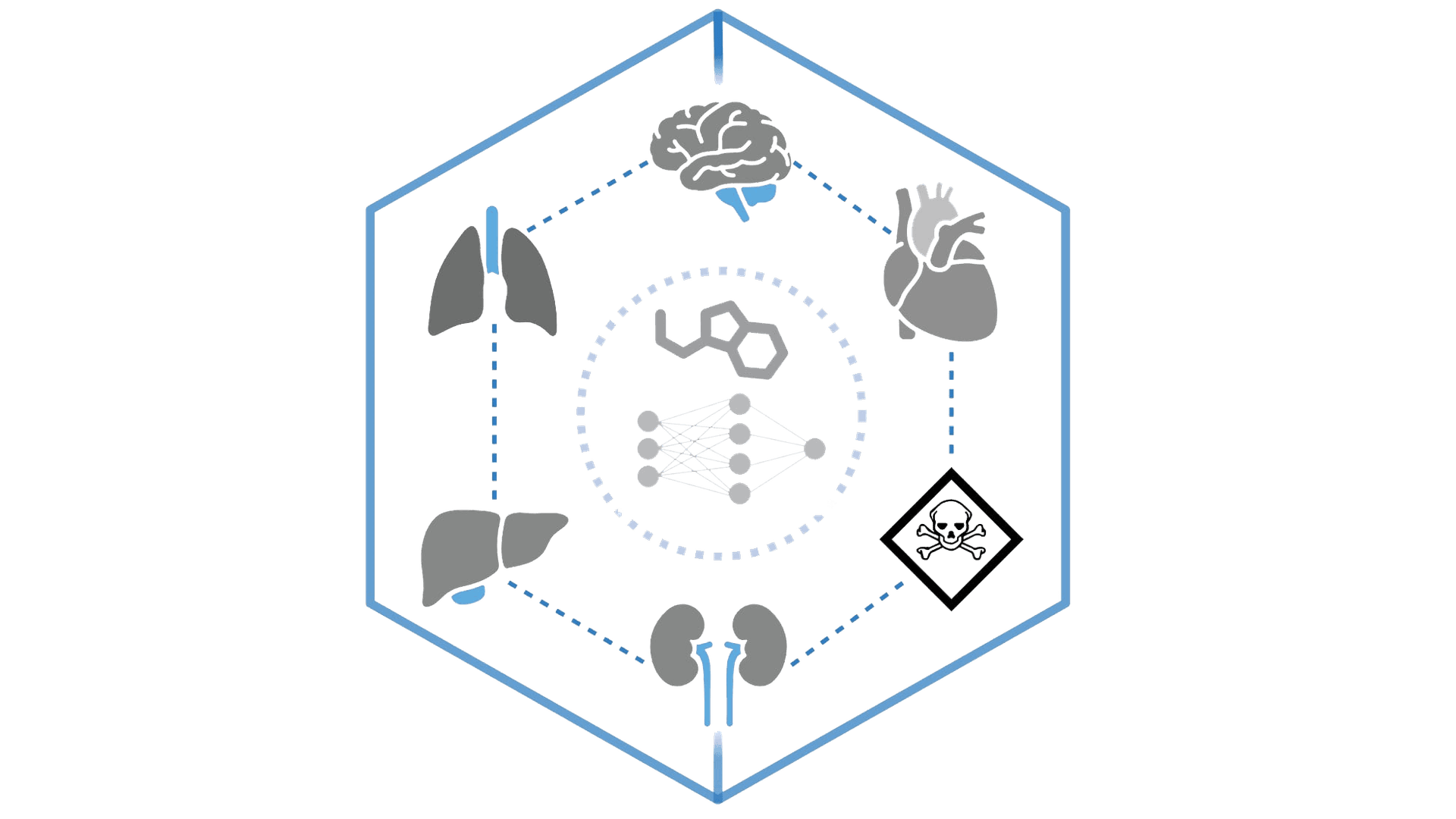
ADMET-AI
Predict absorption, distribution, metabolism, excretion, and toxicity properties using advanced ML for drug development.
eToxPred
Deep learning toxicity prediction for drug candidates. Assess hepatotoxicity, cardiotoxicity, and mutagenicity risks early.
GenMol
AI-powered molecule generation for drug design. Generate novel compounds optimized for target binding and drug-like properties.
PocketFlow
Deep learning pocket detection and druggability assessment. Identify and characterize binding sites on protein targets.

Lipinski's rule of 5
Filter compounds for drug-like properties using molecular weight, lipophilicity, and hydrogen bonding criteria.

Molecular descriptors
Calculate comprehensive molecular properties for QSAR modeling, virtual screening, and lead optimization workflows.
Why researchers choose ProteinIQ
AI-powered ADMET prediction screens hundreds of compounds in seconds
ADMET assays are expensive and time-consuming in the lab
Early toxicity screening identifies red flags before synthesis
Late-stage failures due to unforeseen toxicity issues
Integrated pipeline from ADMET to docking to lead optimization
Fragmented tools require manual data transfer between steps
Professional-grade tools accessible with a free tier
Limited access to expensive commercial drug discovery suites
How it works
Complete drug discovery pipeline with ADMET prediction, toxicity screening, and lead optimization. Accelerate your path from target identification to clinical candidates.
Input compounds
Upload SMILES, SDF, or draw molecules directly
Admet prediction
Screen for absorption, metabolism, and toxicity
Drug-likeness filter
Apply Lipinski rules and molecular property filters
Prioritize leads
Rank compounds and export top candidates
Related solutions
Discover complementary workflows to enhance your research.
Dock molecules with confidence
Screen protein-ligand and protein-protein interactions using AutoDock Vina, DiffDock, GNINA, and LightDock through a unified cloud platform.
Screen millions, discover one
Filter compound libraries at scale with QEPPI, EvoEF2, and PocketFlow to identify promising drug candidates efficiently.
Protein design that actually works
Generate optimized protein sequences and novel binders using LigandMPNN, ProteinMPNN, SolubleMPNN, BoltzGen, and RFdiffusion—state-of-the-art deep learning and diffusion models.