What is SignalP 6.0?
SignalP 6.0 is a machine learning tool developed at the Technical University of Denmark (DTU) for predicting signal peptides in protein sequences. Signal peptides are short amino acid sequences at the N-terminus of proteins that act as molecular "address labels," directing proteins to specific cellular locations or marking them for secretion. Identifying these sequences is essential for studying protein localization, bacterial pathogenesis, and designing recombinant proteins for biotechnology applications.
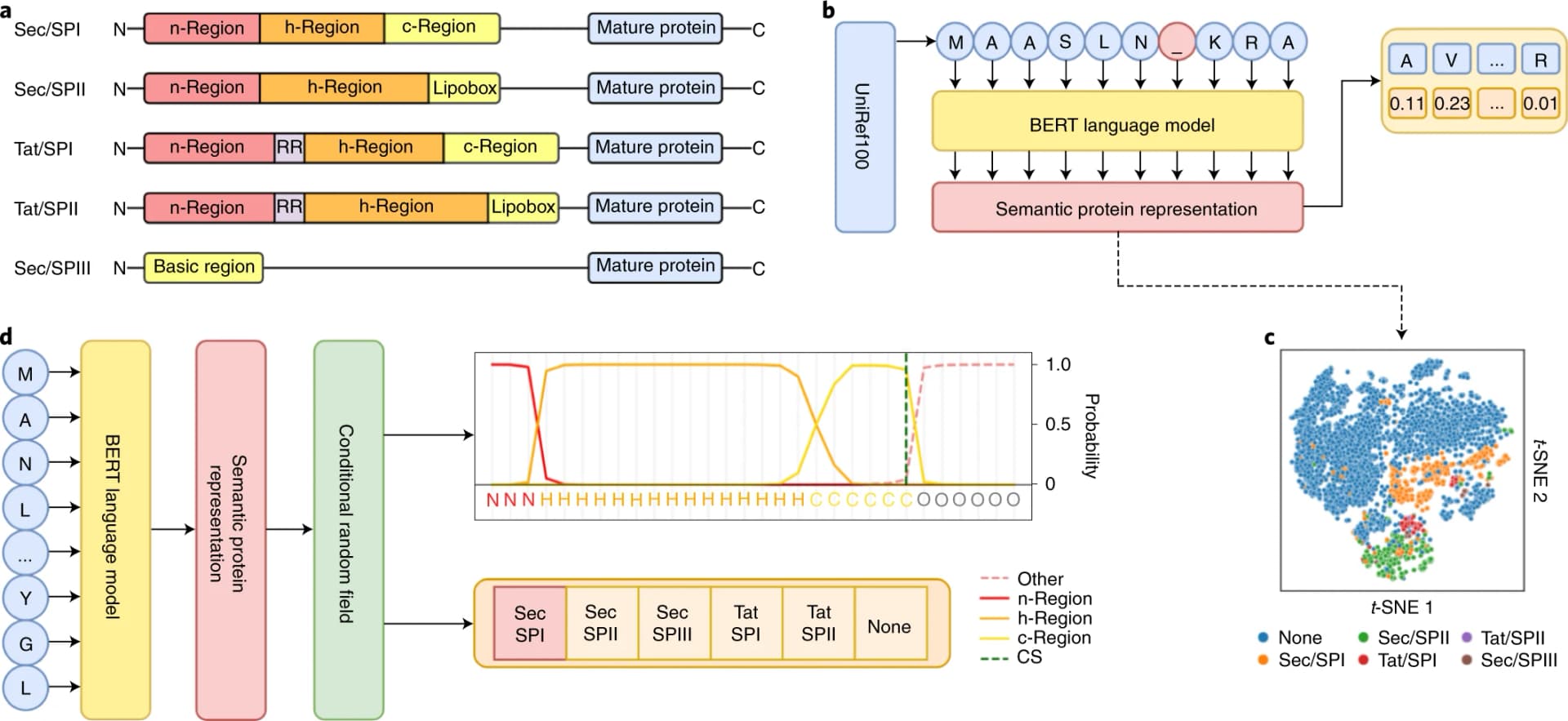
Published in Nature Biotechnology in January 2022, SignalP 6.0 is the first tool capable of detecting all five known types of signal peptides. Previous versions could only reliably detect the first three.
| Type | Secretion Pathway | Peptidase | Characteristics |
|---|---|---|---|
| Sec/SPI | Sec translocon | Signal Peptidase I | Most common type; secreted and cell surface proteins |
| Sec/SPII | Sec translocon | Signal Peptidase II | Prokaryotic lipoproteins; C-terminal lipobox motif |
| Sec/SPIII | Sec translocon | Prepilin peptidase | Type IV pilin-like proteins |
| Tat/SPI | Twin-arginine translocation | Signal Peptidase I | Twin-arginine (RR) motif; folds before transport |
| Tat/SPII | Twin-arginine translocation | Signal Peptidase II | Twin-arginine lipoproteins |
The Sec pathway transports unfolded proteins across membranes, while the Tat (twin-arginine translocation) pathway can transport fully folded proteins.
SignalP has been one of the most widely used bioinformatics tools since the original version was released in 1997. The tool performs best on sequences similar to its training data—novel signal peptide types or highly divergent sequences may be less accurately predicted. Predicted cleavage sites may be off by a few residues, so experimental validation is recommended for critical applications.
How does SignalP 6.0 work?
SignalP 6.0 uses a two-component neural network architecture. The first component is a 30-layer BERT-style transformer model pre-trained on UniRef100, a database containing over 200 million protein sequences. During pre-training, the model learns to predict masked amino acids in protein sequences, similar to how language models learn word relationships in text. This self-supervised training allows the model to capture evolutionary and structural patterns without requiring labeled signal peptide examples.
The second component is a conditional random field (CRF) that takes the language model's representations and predicts whether each position belongs to specific signal peptide regions (n-region, h-region, c-region) or the mature protein, the overall signal peptide type, and the cleavage site position. The CRF ensures predictions follow biologically valid patterns—for example, enforcing that the n-region must precede the h-region.
The model was trained on a curated dataset of experimentally verified signal peptides with strict homology partitioning at 30% sequence identity to prevent data leakage.
| Signal peptide type | Training sequences |
|---|---|
| Sec/SPI | 3,352 |
| Sec/SPII | 2,261 |
| Sec/SPIII | 113 |
| Tat/SPI | 595 |
| Tat/SPII | 36 |
At sequence identities below 60%, SignalP 6.0 outperforms SignalP 5.0. The most notable gains are for Sec/SPIII and Tat signal peptides, which were poorly detected by earlier versions.
How to use SignalP 6.0 online
SignalP 6.0 is available through DTU Health Tech. The service is free but has some constraints: maximum 1,000 proteins per submission, sequences between 10 and 10,000 amino acids, and results remain accessible for 24 hours. The long output format may timeout for more than 100 entries. A mirror on BioLib is available when the primary server is overloaded.
SignalP 6.0 accepts protein sequences in FASTA format. You'll also need to specify the organism group (Eukarya, Gram-positive bacteria, Gram-negative bacteria, or Archaea). The organism group helps the model apply appropriate priors, though SignalP 6.0 can predict signal peptides even without this information, making it suitable for metagenomic sequences of unknown origin.
For local installation, academic users can download the Python package from DTU's website for free. Commercial users must contact DTU's Software Package Manager for licensing. The source code is available on GitHub. The standalone version allows processing larger datasets, integration into pipelines, and offline use.
SignalP 6.0 alternatives
Several other tools can predict signal peptides, including Phobius (which combines signal peptide and transmembrane topology prediction) and PrediSi (specialized for eukaryotic signal peptides).
For understanding the physicochemical properties that make signal peptides functional, analytical tools can provide complementary insights. The hydrophobic H-region is the defining feature of signal peptides, so hydropathy plots can help visualize hydrophobic stretches along a protein sequence. The GRAVY score provides a single value for overall protein hydrophobicity. Tools like Protein Parameters calculate multiple physicochemical properties including molecular weight, isoelectric point, and amino acid composition—useful for characterizing signal peptides and mature proteins separately.
Sources
- Teufel F, et al. "SignalP 6.0 predicts all five types of signal peptides using protein language models." Nature Biotechnology 40, 1023-1025 (2022). doi:10.1038/s41587-021-01156-3
- DTU Health Tech SignalP 6.0 Server
- SignalP 6.0 GitHub Repository


![What's the most common amino acid? [2025 data]](/_next/image?url=%2Fimages%2Fguides%2Fmost-common-amino-acid%2Fhero.png&w=1920&q=75&dpl=dpl_GYaU7FbcykPjH1MLMsWNa7vLwhMa)
