What is the largest protein?
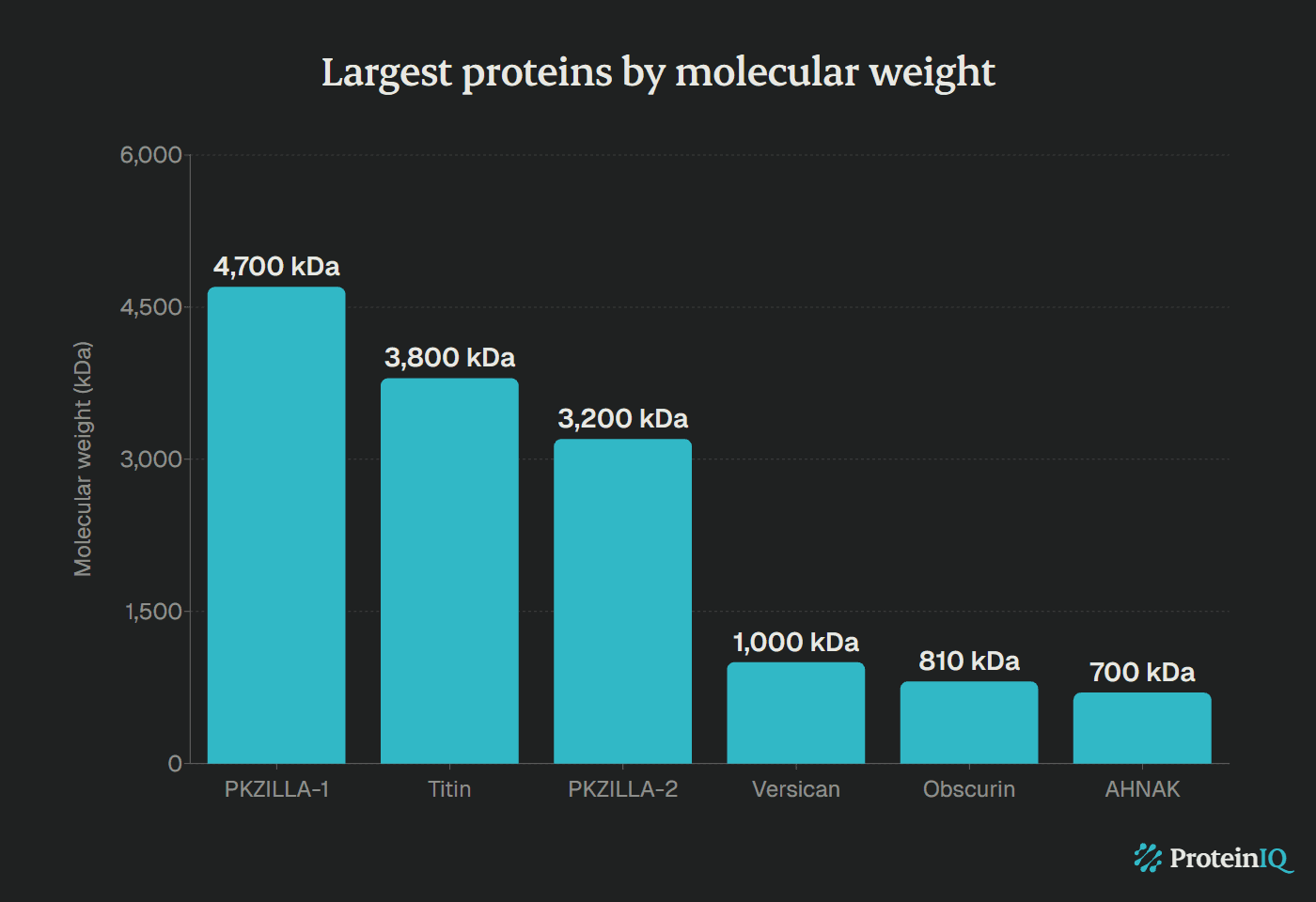
The largest known protein is PKZILLA-1, a giant enzyme containing 45,212 amino acids with a molecular mass of 4.7 megadaltons (MDa). Discovered in August 2024 by researchers at UC San Diego's Scripps Institution of Oceanography, this protein surpasses titin by 25% in molecular weight.
PKZILLA-1 was identified in the golden alga Prymnesium parvum, where it functions as a polyketide synthase enzyme. The protein contains 140 enzyme domains arranged in sequence, making it an extraordinarily complex molecular machine.
"This is the Mount Everest of proteins," noted Bradley Moore, the study's senior author at Scripps Oceanography.
| Feature | PKZILLA-1 | Titin (previous record) |
|---|---|---|
| Mass | 4.7 MDa | 3.7–3.8 MDa |
| Amino acids | 45,212 | 34,350 |
| Enzyme domains | 140 | N/A (structural) |
| Function | Polyketide synthesis | Muscle elasticity |
| Organism | Prymnesium parvum (algae) | Human muscle |
| Discovery | 2024 | 1979 |
How big is titin, the previous record holder?
Titin consists of 34,350 amino acids in its canonical human isoform, with a molecular weight of approximately 3.8 MDa. The protein stretches over 1 micrometer in length, spanning the entire length of the muscle sarcomere from Z-disk to M-line.
Titin functions as a molecular spring in striated muscle, providing passive elasticity that allows muscles to stretch and return to their resting state. According to RCSB PDB-101, titin consists of more than 34,000 amino acids organized into several hundred modular domains, including Ig-like domains, fibronectin-like domains, and a disordered PEVK region.
The mouse homologue is slightly larger, comprising 35,213 amino acids with a molecular weight of 3.9 MDa. Titin constitutes approximately 10% of muscle mass, making it the third most abundant protein in muscle after actin and myosin.
How big is PKZILLA-1 compared to typical proteins?
PKZILLA-1 is approximately 100 times larger than an average protein. According to Bionumbers, eukaryotic proteins average 472 amino acids, while bacterial proteins average 320 amino acids. Using the standard residue weight of 110 Da per amino acid, this translates to approximately 33–55 kDa for a typical protein.
| Protein category | Amino acids | Molecular weight |
|---|---|---|
| Typical protein | 300–500 | 33–55 kDa |
| Large protein | 1,000–5,000 | 100–500 kDa |
| Giant protein (titin) | 34,350 | 3,700–3,800 kDa |
| Largest protein (PKZILLA-1) | 45,212 | 4,700 kDa |
PKZILLA-1 reaches approximately 1 micrometer in length. Despite its enormous size, it remains far smaller than most cells; a typical human cell measures 10–100 micrometers in diameter.
What does PKZILLA-1 do?
PKZILLA-1 and its companion enzyme PKZILLA-2 (3.2 MDa, 99 enzyme domains) work together to produce prymnesin, a potent ichthyotoxin responsible for massive fish kills during harmful algal blooms. According to the Science publication, these giant enzymes catalyze 239 sequential chemical reactions to assemble the complex toxin molecule.
This discovery has two important implications for us:
- Detecting PKZILLA genes in water samples could enable early warning systems for toxic algal blooms. As co-author Timothy Fallon noted, "Monitoring for the genes instead of the toxin could allow us to catch blooms before they start."
- Understanding how nature assembles such complex molecules could inform synthesis of new compounds for medical applications. According to Moore, "Understanding how nature has evolved its chemical wizardry gives us as scientific practitioners the ability to apply those insights to creating useful products."
What are the top 10 largest proteins?
Based on molecular weight, here are the largest known proteins. You can calculate the molecular weight and protein parameters for any sequence using our tools.
| Rank | Protein | Molecular weight | Amino acids | Function |
|---|---|---|---|---|
| 1 | PKZILLA-1 | 4,700 kDa | 45,212 | Polyketide synthesis (algae) |
| 2 | Titin | 3,000–3,800 kDa | 27,000–34,350 | Muscle elasticity |
| 3 | PKZILLA-2 | 3,200 kDa | ~30,000 | Polyketide synthesis (algae) |
| 4 | Versican | ~1,000 kDa | Variable | Extracellular matrix |
| 5 | Obscurin | 720–900 kDa | ~7,000 | Muscle organization |
| 6 | Nebulin | 600–900 kDa | ~6,700 | Thin filament regulation |
| 7 | AHNAK | ~700 kDa | 5,890 | Membrane organization |
| 8 | Ryanodine receptor (RyR1) | ~565 kDa (monomer) | 5,038 | Calcium release channel |
| 9 | Apolipoprotein B-100 | ~515 kDa | 4,536 | Lipid transport |
| 10 | Plectin | ~500 kDa | ~4,500 | Cytoskeletal crosslinking |
Many of the largest proteins are found in muscle tissue (titin, obscurin, nebulin, dystrophin), reflecting the complex structural requirements of the contractile apparatus.
Why are some proteins so large?
Giant proteins serve specialized functions that require their extraordinary size:
- Titin spans the entire muscle sarcomere (~1 μm), physically connecting the Z-disk to the M-line. This requires a protein large enough to bridge this distance while maintaining elastic properties.
- PKZILLA-1 contains 140 enzyme domains that work in sequence. Rather than relying on 140 separate enzymes finding each other through diffusion, consolidating them into a single protein ensures efficient, sequential processing of chemical reactions.
- Large muscle proteins like titin, nebulin, and obscurin act as molecular rulers that define sarcomere dimensions and organize other proteins during muscle development.
Jon Clardy, a biological chemist at Harvard, noted that these PKZILLA proteins likely approach the upper size limits for proteins because of the sheer workload of expressing and assembling something that large.
How many different proteins exist?
While PKZILLA-1 represents the extreme upper end of protein size, the total number of known protein sequences exceeds 465 million (UniRef100, 2025). The human genome encodes approximately 19,000–20,000 protein-coding genes, which produce roughly 70,000 distinct protein isoforms through alternative splicing.


![What's the most common amino acid? [2025 data]](/_next/image?url=%2Fimages%2Fguides%2Fmost-common-amino-acid%2Fhero.png&w=1920&q=75&dpl=dpl_GYaU7FbcykPjH1MLMsWNa7vLwhMa)

![How many proteins are there? [2026 data]](/_next/image?url=%2Fimages%2Fguides%2Fnumber-of-proteins%2Fhero.jpg&w=1920&q=75&dpl=dpl_GYaU7FbcykPjH1MLMsWNa7vLwhMa)