We've integrated BoltzGen, a state-of-the-art AI model from MIT for designing protein and peptide binders against any biomolecular target. Released in 2025 by Hannes Stärk and the MIT Jameel Clinic, BoltzGen uses generative diffusion models—the same architecture that powers AI image generators—to create entirely novel binders from scratch.
Binder design is foundational for drug discovery, therapeutic development, and basic research. Whether you're designing peptide therapeutics, miniprotein inhibitors, or nanobody-based biologics, understanding how to create molecules that bind specifically to your target drives everything from lead generation to functional validation. But designing binders has traditionally required deep computational expertise, expensive infrastructure, and weeks of iterative development.
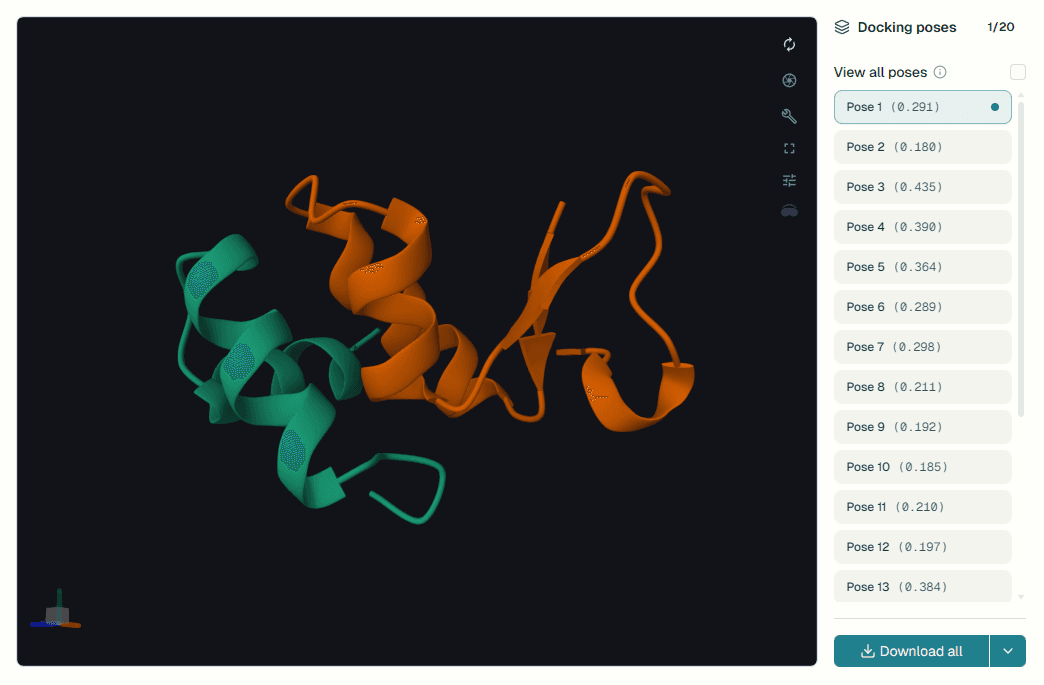
ProteinIQ makes BoltzGen easily accessible, allowing you to upload a target structure, configure your design parameters, and get ranked binder candidates with quality predictions in minutes.
Universal binder design with all-atom precision
BoltzGen represents a paradigm shift in computational protein design. Unlike traditional methods that modify existing sequences or work at reduced resolution, BoltzGen generates entirely novel binders through an all-atom diffusion process that models every heavy atom with precise 3D geometry. The model employs an innovative geometry-based residue representation where each designed position is encoded as 14 virtual atoms—four for the backbone (N, C-α, C, O) and ten additional atoms whose 3D positions geometrically encode amino acid identity. This elegant approach enables scalable joint training on both structure prediction and design tasks without the computational challenges of mixing discrete sequence identities with continuous coordinates.
The model can design peptides ranging from 8 to 30 residues for small binding pockets and surface grooves, proteins from 50 to 150 residues for complex protein-protein interfaces, and nanobodies of 110 to 130 residues optimized for therapeutic applications. It works against diverse targets, including proteins, nucleic acids, small molecules, and even intrinsically disordered regions—dramatically expanding the druggable universe beyond traditional therapeutic targets.
BoltzGen's architecture is built upon AlphaFold3 and Boltz-2, comprising a large Trunk (backbone network) and a Diffusion Module. During training, the model randomly performs structure prediction, binder design, and structure completion tasks on each iteration. This multi-task learning enables BoltzGen to leverage structural knowledge from the entire PDB for design tasks, while design data improves structure prediction capabilities, simultaneously achieving state-of-the-art performance on both generative design and structure prediction.
Our implementation runs on NVIDIA A100 GPUs with optimized infrastructure. Peptide designs are complete in 5-15 minutes, while protein designs take 15-30 minutes. The model generates multiple candidates with confidence scores, enabling rapid iteration during lead discovery.
Why BoltzGen
We chose BoltzGen because it achieves unprecedented experimental validation rates while maintaining the flexibility to design against any target type. In rigorous wet-lab testing across 26 distinct targets, BoltzGen achieved a 66% overall success rate with nanomolar or better binding affinity—testing only 15 or fewer designs per target.
On benchmark targets with known binding structures, such as PD-L1, TNFα, and PDGFR, BoltzGen achieved an 80% success rate for both nanobodies and proteins, including picomolar hits. On a challenging set of novel targets with less than 30% sequence similarity to training data, the model delivered nanomolar binders for 6 out of 9 targets, with best Kd values reaching 7.8 nM, 6.1 nM, and 8.8 nM.
The all-atom diffusion approach provides several advantages over physics-based or sequence-based methods. It generates entirely novel sequences, not constrained by existing scaffolds, and works across diverse target types without requiring retraining. BoltzGen introduces a flexible design specification language that provides precise control over the generation process through multiple constraint types simultaneously: covalent bonds for cyclic peptides or disulfide-stapled designs, structure conditioning via pairwise distance constraints, binding site targeting for specific target residues, secondary structure constraints (α-helices, β-sheets, coils), and design masks controlling which positions can be designed versus fixed. These constraints guide the diffusion process during inference toward specific design objectives, eliminating the need for model retraining.
The computational pipeline consists of distinct steps that vary depending on the protocol. All protocols begin with diffusion (generating 3D atomic coordinates) and inverse folding (converting structures to amino acid sequences). The protein-anything protocol introduces a refolding step to verify structural stability, resulting in a 4.9-fold increase in computational cost compared to baseline peptides. The protein-small_molecule protocol includes both refolding and affinity prediction steps for a 6.8× cost multiplier, making it the most computationally intensive modality.
The tool accepts target structures as PDB/CIF files or RCSB PDB IDs, along with an optional binding site specification for focused design. You can configure the binder type with different cost profiles (peptides and nanobodies at 1.0× baseline, proteins at 4.9×, and protein-small_molecule at 6.8×). The flexible size range spans 8-500 residues, and the cyclic topology provides enhanced peptide stability. Each design includes ranked candidates with quality metrics like pTM score (predicted structural quality), PAE (prediction uncertainty in Ångströms), and buried surface area (interface size), complete 3D structure coordinates in CIF format, designed amino acid sequences in FASTA format, interactive 3D visualization in the browser, and downloadable files for further validation.
Getting started
BoltzGen is available now on ProteinIQ to everyone.
For typical usage, upload or fetch your target structure in PDB/CIF format or by PDB ID, choose your binder type and size, optionally specify the binding site if known from literature, configure the number of designs (10-20 for exploration, 100+ for screening) and budget parameter (typically equal to number of designs for receiving all candidates, or lower for diversity-optimized subsets), then submit and review ranked results in approximately 5-30 minutes depending on design complexity.
Begin with peptides for proof-of-concept work, as they're fast and inexpensive. The peptide-anything protocol runs at baseline cost (1.0× multiplier) and completes quickly, making it ideal for initial validation. Nanobodies also run at baseline cost despite longer sequences (110-130 residues). When you have experimental evidence about binding sites, specify them to improve the success rate and reduce computational cost by focusing the generation on known functional regions. Generate 20-40 designs for initial exploration to sample a diverse range of binding modes. Clean your target structures using PDB Fixer before submission to ensure optimal model performance. Most importantly, validate the top designs experimentally.
The model generalizes well to unseen targets and interaction motifs. BoltzGen's 66% experimental success rate means testing multiple top-ranked candidates significantly increases your probability of finding successful binders.
Further information
BoltzGen was developed by Hannes Stärk and the MIT Jameel Clinic for Machine Learning in Health. The model is based on research published in "BoltzGen: Toward Universal Binder Design" (2025). The architecture builds on AlphaFold3 and Boltz-2, employing all-atom generative diffusion with geometry-based residue representation for joint structure prediction and design.
For technical details, see the BoltzGen tool page. For questions about infrastructure, pricing, or custom deployments, contact our team.
Every researcher deserves access to the tools that accelerate discovery. We're here to make that happen
